Quantitative microbial ecology through stable isotope probing
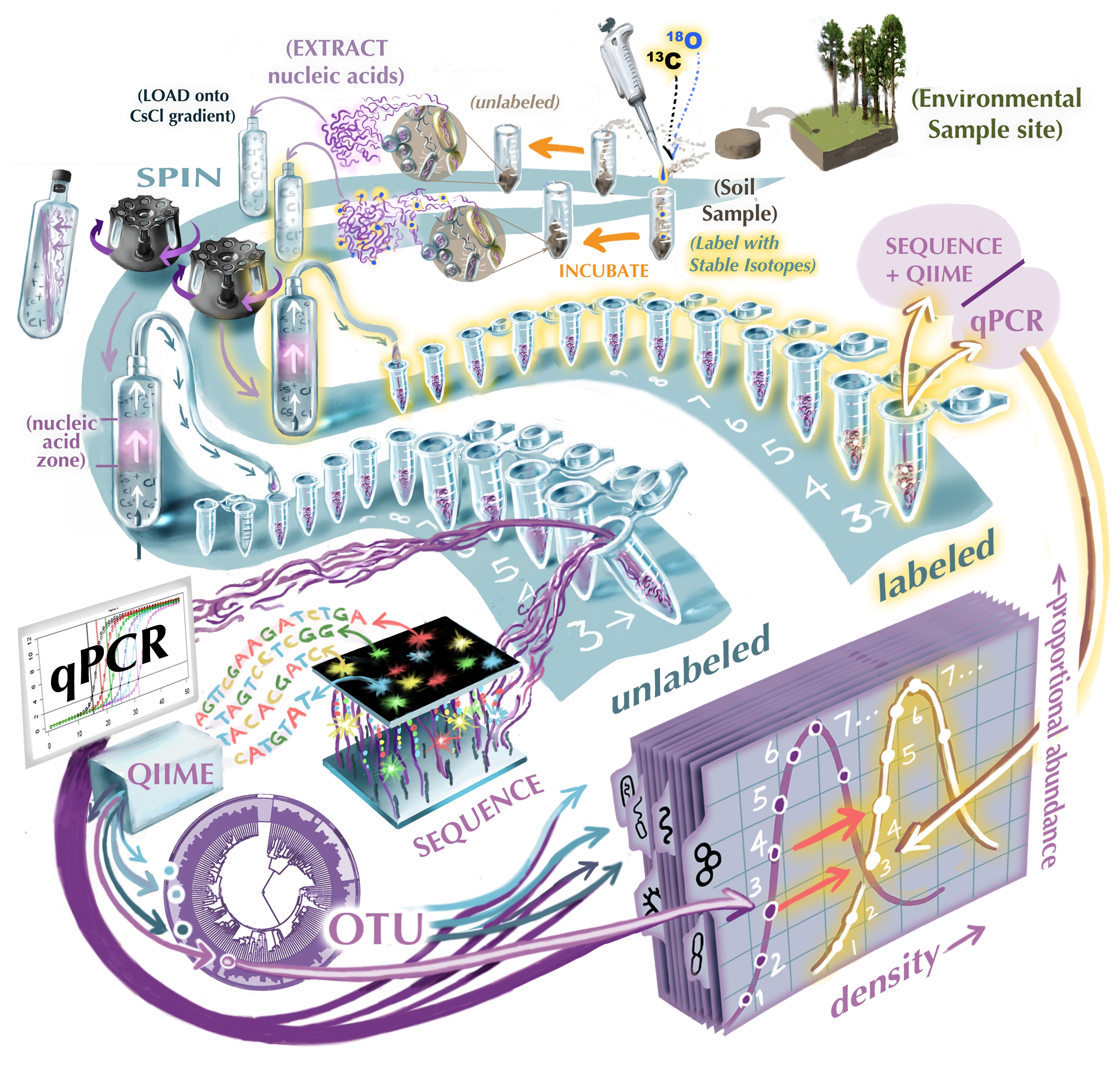
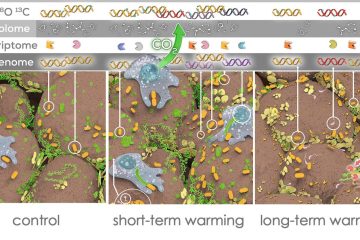
 Measuring the growth rates of individual bacteria taxa within a mixed sample gives a powerful view of the moving parts of a living ecosystem, especially when that sample is soil, where so many ecological processes occur. Ecoss research pioneered “qSIP” — a new method for gathering many simultaneous bacterial growth curves from an environmental sample by combining Stable Isotope Labeling with next-generation sequencing techniques: as shown in the illustration, stable isotope labels are added to a sample and incorporated into the DNA of growing bacteria. Fractionating the sample by density allows the density of each taxon in the sample to be identified separately by combining qPCR with 16s rRNA sequencing (shown here as individual curves plotted over each taxon’s appearance in the density column). Finally, comparing the difference in density for each taxon between labeled and unlabeled halves of the incubated sample reveals the rate of isotope assimilation, and thereby each taxon’s rate of growth.
Measuring the growth rates of individual bacteria taxa within a mixed sample gives a powerful view of the moving parts of a living ecosystem, especially when that sample is soil, where so many ecological processes occur. Ecoss research pioneered “qSIP” — a new method for gathering many simultaneous bacterial growth curves from an environmental sample by combining Stable Isotope Labeling with next-generation sequencing techniques: as shown in the illustration, stable isotope labels are added to a sample and incorporated into the DNA of growing bacteria. Fractionating the sample by density allows the density of each taxon in the sample to be identified separately by combining qPCR with 16s rRNA sequencing (shown here as individual curves plotted over each taxon’s appearance in the density column). Finally, comparing the difference in density for each taxon between labeled and unlabeled halves of the incubated sample reveals the rate of isotope assimilation, and thereby each taxon’s rate of growth.