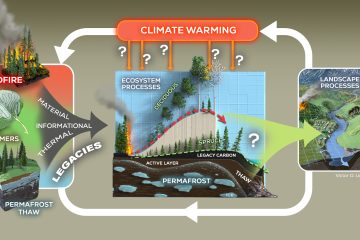
mRNA, rRNA and DNA quantitative stable isotope probing with H218O indicates use of old rRNA among soil Thaumarchaeota
RNA is considered to be a short-lived molecule, indicative of cellular metabolic activity, whereas DNA is thought to turn over more slowly because living cells do not always grow and divide. To explore differences in the rates of synthesis of these nucleic acids, we used H218O quantitative stable isotope probing (qSIP) to measure the incorporation of 18O into 16S rRNA, the 16S rDNA, amoA mRNA and the amoA gene of soil Thaumarchaeota. Incorporation of 18O into the thaumarchaeal amoA mRNA pool was faster than into the 16S rRNA pool, suggesting that Thaumarchaea were metabolically active while using rRNA molecules that were likely synthetized prior to H218O addition. Assimilation rates of 18O into 16S rDNA and amoA genes were similar, which was expected because both genes are present in the same thaumarchaeal genome. The Thaumarchaea had significantly higher rRNA to rDNA ratios than bacteria, though the 18O isotopic signature of thaumarchaeal rRNA was lower than that of bacterial rRNA, further suggesting preservation of old non-labeled rRNA. Through qSIP of soil with H218O, we showed that 18O incorporation into thaumarchaeal nucleic acids was generally low, indicating slower turnover rates compared to bacteria, and potentially suggesting thaumarchaeal capability for preservation and efficient reuse of biomolecules.