Modeling soil metabolic processes using isotopologue pairs of position-specific 13 C-labeled glucose and pyruvate
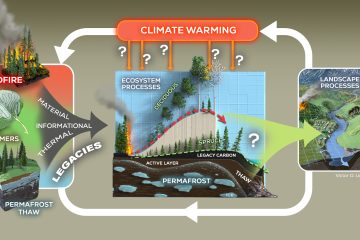
Most organic carbon (C) in soils eventually turns into CO2 after passing through microbial metabolic pathways, while providing cells with energy and biosynthetic precursors. Therefore, detailed insight into these metabolic processes may help elucidate mechanisms of soil C cycling processes. Here, we describe a modeling approach to quantify the C flux through metabolic pathways by adding 1-13C and 2,3-13C pyruvate and 1-13C and U-13C glucose as metabolic tracers to intact soil microbial communities. The model calculates, assuming steady-state conditions and glucose as the only substrate, the reaction rates through glycolysis, Krebs cycle, pentose phosphate pathway, anaplerotic activity through pyruvate carboxylase, and various biosynthesis reactions. The model assumes a known and constant microbial proportional precursor demand, estimated from literature data. The model is parameterized with experimentally determined ratios of 13CO2 production from pyruvate and glucose isotopologue pairs. Model sensitivity analysis shows that metabolic flux patterns are especially responsive to changes in experimentally determined 13CO2 ratios from pyruvate and glucose. Calculated fluxes are far less sensitive to assumptions concerning microbial chemical and community composition. The calculated metabolic flux pattern for a young volcanic soil indicates significant pentose phosphate pathway activity in excess of pentose precursor demand and significant anaplerotic activity. These C flux patterns can be used to calculate C use efficiency, energy production and consumption for growth and maintenance purposes, substrate consumption, nitrogen demand, oxygen consumption, and microbial C isotope composition. The metabolic labeling and modeling methods may improve our ability to study the biochemistry and ecophysiology of intact and undisturbed soil microbial communities.