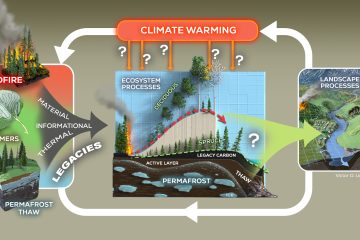
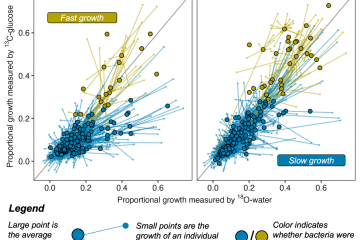
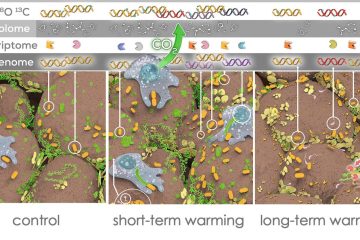
Multiple-Element Isotope Probes, NanoSIMS, and the Functional Genomics of Microbial Carbon Cycling in Soils in Response to Chronic Climatic Change
For the past several decades, connecting biogeochemistry and microbial genomics has been a high priority in microbial ecology. Yet, techniques that actually link element flow and genomic information are scarce. In this project, we are using the Chip-SIP method to measure isotopic composition of major elements (C, N, H, and O) of nucleic acid sequences representing individual microbial taxa. RNA is extracted from an environmental sample after exposure to isotopically labeled substrates. The nucleic acids from the entire microbial community are then exposed to a microarray containing small probes that target the 16S rRNA genes of a large variety of microorganisms so that nucleic acids extracted from the environmental sample bind to matching probes. Then, the entire microarray is placed under a nanoscale secondary ion mass spectrometer, which sequentially analyzes the RNA bound to each probe for isotopic composition. In this way, element flow in the natural environment into individual microbial taxa can be determined.