New tool reveals role of ancestry in soil communities of bacteria
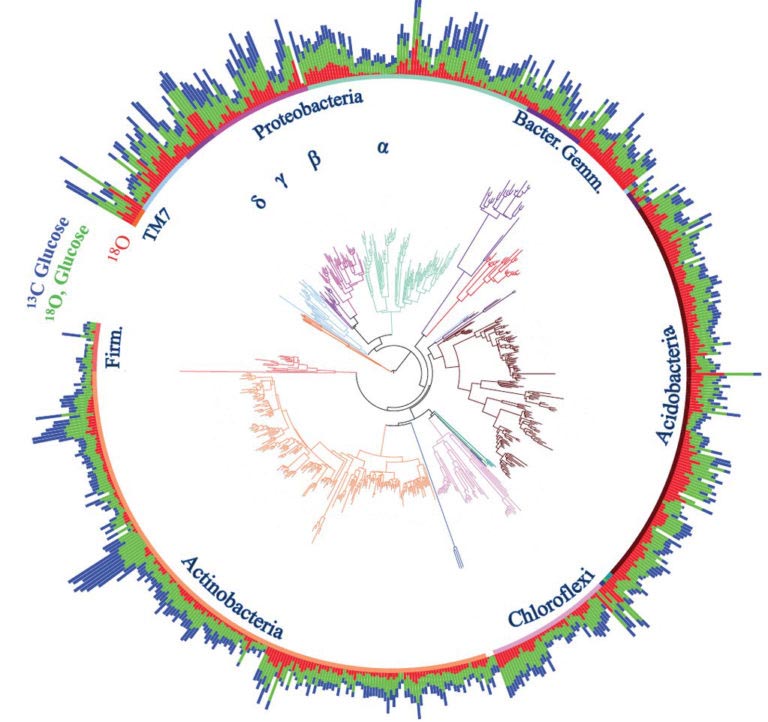
Clustering in the use of carbon and oxygen shows an influence of evolutionary history on bacterial activity in soil. (Image courtesy Morrissey, et al. 2016.)
The link between lineage and behavior has inspired research across the spectrum of life. For plants and animals, genetically close cousins tend to act in similar ways. Finches, for example, eat seeds, while swallows eat insects. For bacteria, however, the question is up for debate.
Does evolutionary history predict how different types of bacteria behave and function? Or, conversely, do unrelated bacteria typically overlap each other’s functions, providing the same ecosystem services—a concept known as functional redundancy? The answer determines how much may be lost when strains of bacteria are lost from a soil community.
Ecoss researchers at Northern Arizona University led by post-doctoral researcher Ember Morrissey—now an assistant professor of Environmental Microbiology at West Virginia University—took a close-up look at differences in bacterial activity in intact soil communities using a new tool. Quantitative stable isotope probing, known as qSIP, is an improvement over existing qualitative methods because it measures more than just the presence or absence of certain types of bacterial activity in soil—it measures the rate of that activity. Activity rate can then be linked to the DNA of a specific bacterial strain to look for patterns of behavior at a much finer scale.
“Using this new approach,” said Morrissey, “we can now answer many questions about the ecology of microorganisms and their role in determining ecosystem function that were previously out of reach.”
The research was funded by a $1.4 million grant from the Department of Energy, Genomic Sciences Program. Results were recently published in The ISME Journal.
Researchers incubated soil samples from an intact Northern Arizona ponderosa pine forest with water and glucose that were uniquely marked. The water contained a heavier form (isotope) of oxygen, and the glucose contained a heavier form of carbon. The amount of isotope used by different types of bacteria, measured using qSIP, is like a meter that shows how fast the bacteria grow and how much carbon they use.
Bacteria sharing a closer evolutionary history showed similar population growth rates and carbon use. Bacteria farther apart by lineage differed in these behaviors. The rates that different types of bacteria grow and assimilate carbon both affect how an ecosystem functions. This suggests that different types of bacteria may not always be able to substitute for each other’s contribution to important processes.
Evolutionary history, or phylogeny, it turns out, does play a role in the activity of bacteria in intact soil communities. Linking bacterial lineages to specific types of activity in the soil offers a new way to understand how the specific mix of bacteria in soil influences ecosystem function.
Sonya Daw
news.nau.edu