Linking Biogeochemistry and Microbial Community Chemistry
 Overview
Overview
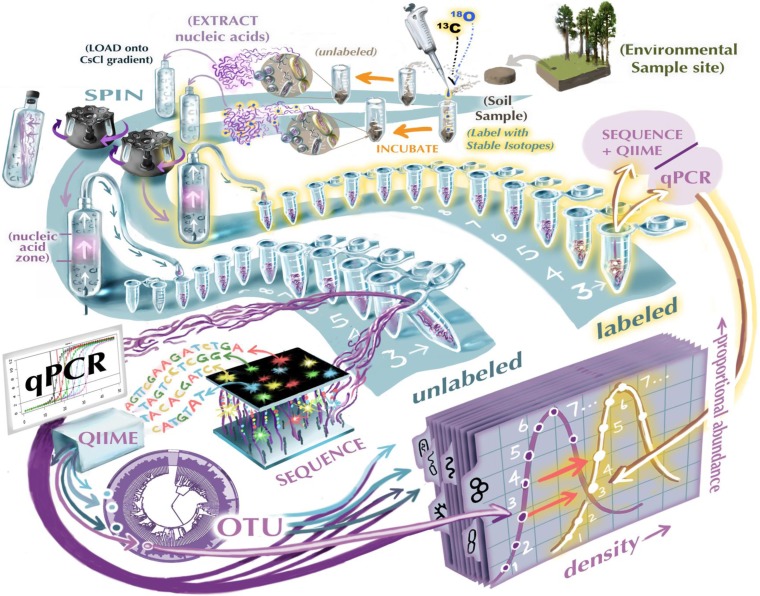
The identities of organisms profoundly influence ecosystems, and microbial diversity is vast. The obvious disconnect between this diversity and its treatment in C cycling models is perhaps the best manifestation of the common complaint that so much physiology, taxonomy, ecology, and diversity is ignored when microbial communities are described with boxes and arrows. A recent report from the American Society for Microbiology captured this complaint by depicting on the cover the hackneyed “black box” of microbial ecology, although the report emphasized the promise of new approaches by showing the box open and illuminated from within. In this project, we are exploring relationships between the diversity of soil microorganisms and the processing of soil carbon and its conversion from organic C to carbon dioxide (CO2). Our approach is designed to connect multiple element biogeochemistry and microbial community ecology, by linking isotopes to genomics, taking advantage of developments in stable isotope probing and isopycnic separation, microarrays and sequencing technology, and NanoSIMS.
Details
Tackling this problem is important to understanding soil C persistence, because the sensitivity of soil C to external forcings has the potential to mitigate or exacerbate global climate change. Warming can cause soil C release, and these losses have implications for the distribution and retention of nitrogen, which in turn can feed back to the C cycle through changes in plant productivity. Increased atmospheric carbon dioxide (CO2) increases photosynthesis but not necessarily soil C, and in some cases can even cause soil C loss. Soil C content changes with long-term differences in precipitation, and soil CO2 production exhibits strong short-term changes in response to wet-dry cycles. Multiple global environmental changes drive changes in soil C, but patterns in the diversity of organism driving these changes are poorly understood. Which organisms catalyze soil C losses? Are the same organisms involved when the forcings altering soil C are different? Are the activities of the organisms related to their phylogenies? What are their growth rates, and relative reliance on more labile C sources, such as those derived from rhizoexudates by living plants, or from plant litter? How do their activities interact with other resources, like nutrients? These are broad questions that to date have been nearly impossible to answer on a scale appropriate for the diversity of soils, because of the difficulty of coupling biogeochemistry with genomics. Understanding which microorganisms catalyze soil C losses is especially difficult, because the chemical forms of native soil organic C are diverse, heterogeneous, and not well characterized, making it impractical to identify the organisms that utilize them using classical stable isotope probing. We have developed advanced SIP methods that use isotope tracers to quantify the growth (18O, 2H) and carbon (13C) and nitrogen (15N) sources of specific microorganisms that are actively degrading soil organic C. We will also use secondary ion mass spectrometry coupled with microarrays to allow high-level taxonomic resolution.
Related publications
Mau RL, Liu CM, Aziz M, Schwartz E, Dijkstra P, Marks JC, Price LB, Keim P, Hungate BA, 2015. Linking soil bacterial biodiversity and soil carbon stability. ISME 9, 1477-1480.
Grants supporting this work
- Department of Energy, Systems Biology Enabled Research on the Role of Microbial Communities in Carbon Cycling: Multiple element isotope probes, NanoSIMS, and the functional genomics of microbial carbon cycling in soils in response to chronic climatic change, $1,430,493, 9/13-8/16
Research topics covered by funding source
- Ecosystem Sciences and Metagenomics: The Role of Microorganisms in Soil Carbon Losses
- Future of the Soil Carbon Sink: the Priming Effect
- Taxonomic, genomic, and functional diversity of soil carbon dynamics


