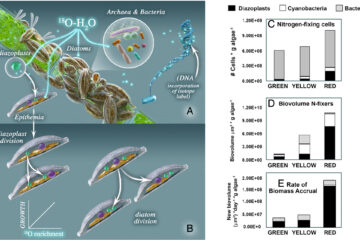
13C analysis of fatty acid fragments by gas chromatography mass spectrometry for metabolic flux analysis
When multiple metabolic pathways lead to the same product, compound-specific isotope analysis may not provide enough information to quantify the activities of the contributing pathways. Instead, identification of where in the molecule the 13C is incorporated is required. Here we show how knowledge of position-specific 13C incorporation in fatty acids (FA) and FA fragments can be used to quantitatively estimate the fluxes through the central C metabolic network. We developed a method to measure 13C enrichment of FA and FA fragments (ethanoate, propionate) using electron impact GC–MS. We tested the accuracy and repeatability of the measurements using natural abundance and position-specific 13C labelled standards and FA extracted from Bacillus licheniformis and Pseudomonas fluorescens grown with labelled and unlabelled glucose. The molecular ions of FA generally reflected theoretical predictions of mass isotopomer distributions for natural abundance values, but that of the associated FA fragments deviated from expected values, likely associated with McLafferty rearrangements of hydrogen. After correction for naturally occurring isotopes, 13C enrichments of FA and FA fragments showed good agreement with expected isotope composition of FA standards (root mean square error < 0.044 at%; δ13C of ∼ 40‰), natural abundance and labelled glucose. The unsaturated FA extracted from P. fluorescens deviated from expected values likely associated with problems of co-elution and ion suppression and were excluded from analysis. The ratio of glucose-1-13C to glucose-3-13C incorporation into FA fragments was high for B. licheniformis, but low for P. fluorescens. Metabolic flux modelling based on the 13C enrichment of ethanoate and propionate fragments showed that B. licheniformis used Embden-Meyerhof-Parnas and pentose phosphate pathway (66% and 30%, respectively), whereas P. fluorescens utilized Entner-Doudoroff and pentose phosphate pathway (72% and 27%, respectively). FA fragment analysis is therefore a promising tool to study central C metabolic network activities of co-occurring groups of microbes in intact and complex environmental communities.