Identification of growing bacteria during litter decomposition in freshwater through H2 18O quantitative stable isotope probing
Identification of microorganisms that facilitate the cycling of nutrients in freshwater is paramount to understanding how these ecosystems function. Here, we identify growing aquatic bacteria using  quantitative stable isotope probing. During 8 day incubations in 97 atom %
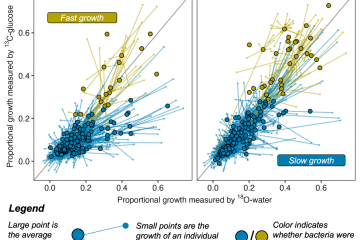
quantitative stable isotope probing. During 8 day incubations in 97 atom % , 54% of the taxa grew. The most abundant phyla among growing taxa were Proteobacteria (45%), Bacteroidetes (30%) and Firmicutes (10%). Taxa differed in isotopic enrichment, reflecting variation in DNA replication of bacterial populations. At the class level, the highest atom fraction excess was observed for OPB41 and δ-Proteobacteria. There was no linear relationship between 18O incorporation and abundance of taxa. δ-Proteobacteria and OPB41 were not abundant, yet the DNA of both taxa was highly enriched in 18O. Bacteriodetes, in contrast, were abundant but not highly enriched. Our study shows that a large proportion of the bacterial taxa found on decomposing leaf litter grew slowly, and several low abundance taxa were highly enriched. These findings indicating that rare organisms may be important for the decomposition of leaf litter in streams, and that quantitative stable isotope probing with
, 54% of the taxa grew. The most abundant phyla among growing taxa were Proteobacteria (45%), Bacteroidetes (30%) and Firmicutes (10%). Taxa differed in isotopic enrichment, reflecting variation in DNA replication of bacterial populations. At the class level, the highest atom fraction excess was observed for OPB41 and δ-Proteobacteria. There was no linear relationship between 18O incorporation and abundance of taxa. δ-Proteobacteria and OPB41 were not abundant, yet the DNA of both taxa was highly enriched in 18O. Bacteriodetes, in contrast, were abundant but not highly enriched. Our study shows that a large proportion of the bacterial taxa found on decomposing leaf litter grew slowly, and several low abundance taxa were highly enriched. These findings indicating that rare organisms may be important for the decomposition of leaf litter in streams, and that quantitative stable isotope probing with  can be used to advance our understanding of microorganisms in freshwater by identifying species that are growing in complex communities.
can be used to advance our understanding of microorganisms in freshwater by identifying species that are growing in complex communities.