Genetic and environmental controls of microbial communities on leaf litter in streams
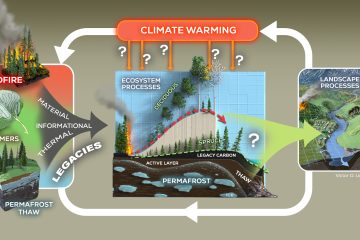
1. Despite the importance of microorganisms for leaf litter decomposition in streams, little is known about which factors affect community composition of bacterial and fungal communities. Standard morphological techniques probably underestimate microbial diversity.
2. We used terminal restriction fragment length polymorphisms of the ITS regions for fungi, and the 16S region for bacteria, to compare fungal and bacterial communities on four cross types of cottonwood leaves (Populus fremontii, P. angustifolia, and their naturally occurring F1 and backcross hybrids). Decomposing leaves were studied in two Arizona rivers that differ in water chemistry and macroinvertebrates.
3. Hybridising cottonwoods are an ideal model system to test how genetic differences in leaf litter chemistry affect microbial communities because cross types have different decomposition rates and leaf litter chemistry. Leaves were incubated in litter bags for
2 weeks and brought to the laboratory for genetic analysis. Communities were analysed using non-metric multi dimensional scaling (NMDS) and diversity indices.
4. Fungal and bacterial communities differed between the two rivers, even when growing on identical substrates. There were also significant differences in microbial communities among the four cross types, indicating that genetically based differences in leaf litter translate to differences in microbial communities.
5. Diversity increased along the hybridising complex from P. fremontii to P. angustifolia, with hybrids showing intermediate values. Fungal and bacterial diversity were significantly higher on cross types with higher tannin concentrations and slower decomposition rates. 6. Environmental conditions most strongly structured microbial communities, but within an environment, genetic-based differences in leaf litter quality yielded differences in diversity and community structure.
7. Molecular tools are making it possible to understand patterns of microbial diversity in river ecosystems, paving the way for a better understanding of how differences in microbial species affect ecosystem processes and higher trophic levels.